※ PhosSNP INTRODUCTION:
As we are entering the age of "Personal Genomics" or "Personalized Medicine", it has been expected that the knowledge of human genetic polymorphisms and variations could provide a foundation for understanding differences in susceptibility to diseases and designing individualized therapeutic treatments (Cargill, et al., 1999; Collins, et al., 1998). Recent progresses of the International HapMap Project and similar projects (International HapMap Consortium, 2005; Frazer, et al., 2007) have provided a wealth of information detailing tens of millions human genetic variations between individuals, including copy number variations (CNVs) (Redon, et al., 2006) and single nucleotide polymorphisms (SNPs) (Hinds, et al., 2005). It was estimated that ~90% of human genetic variations are due to SNPs (Collins, et al., 1998). In particular, by changing amino acids in proteins, non-synonymous SNPs (nsSNPs) in the gene coding regions could account for nearly half of the known genetic variations linked to human inherited diseases (Stenson, et al., 2003). In this regard, numerous efforts have been contributed to elucidate how nsSNPs generate deleterious effects on the stability and function of proteins. Obviously, an nsSNP might change the physicochemical property of a wild-type amino acid to affect the protein stability and dynamics, or disrupt the interacting interface that prohibits the protein to form a complex with its partners (Kono, et al., 2008; Stitziel, et al., 2004; Uzun, et al., 2007; Yue and Moult, 2006). Alternatively, nsSNPs could also influence post-translational modifications (PTMs) of proteins (eg., phosphorylation), by changing the residue types of the target sites or key flanking amino acids (Erxleben, et al., 2006; Gentile, et al., 2008; Ryu, et al., 2009; Savas and Ozcelik, 2005; Yang, et al., 2008). Previously, the Armstrong group firstly coined the term of phosphorylopathy to describe human genetic variation that results in aberrant regulation of protein phosphorylation (Erxleben, et al., 2006; Gentile, et al., 2008).
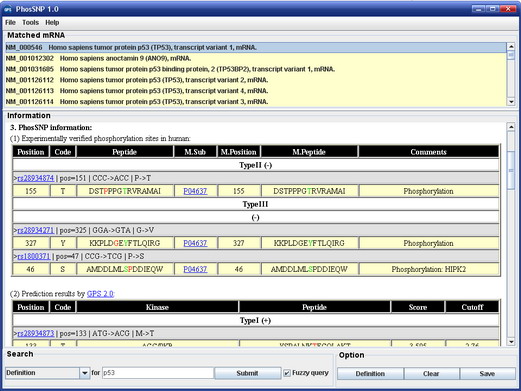
In this work, we performed a genome-wide analysis of genetic polymorphisms that influence protein phosphorylation in H. Sapiens. We collected 91,797 nsSNPs from NCBI dbSNP build 130 (Sherry, et al., 2001). The human mRNA/protein sequences were taken from RefSeq build 31 (Pruitt, et al., 2007). We used our GPS 2.0 software (Xue, et al., 2008) to predict kinase-specific phosphorylation sites for human proteins and nsSNP data. For simplicity, we defined a phosSNP (Phosphorylation-related SNP) as an nsSNP that might influence protein phosphorylation status. We classified all phosSNPs into five groups. The first three types (I, II, and III) were similarly defined as previously described (Ryu, et al., 2009), including change of an amino acid with S/T/Y residue or vice versa to create a new [Type I (+)] or remove an original phosphorylation site [Type I (-)], variations to add [Type II (+)] or remove adjacent phosphorylation sites [Type II (-)], and mutations to change PK types of adjacent phosphorylation sites (Type III) (Ryu, et al., 2009). Also, we observed that an amino acid substitution among S, T or Y could also change the PK types in the phosphorylated position (Type IV), say, the target site could still be phosphorylated but by a different type of kinase. Moreover, we defined the type V phosSNP as a variation that results in a stop codon, which might remove its following phosphorylation sites in the protein C-terminus. Unexpectedly, we computationally detected 69.76% of nsSNPs as potential phosSNPs (64, 035) in 17, 614 proteins. In this regard, we proposed that most of nsSNPs might affect protein phosphorylation and play ubiquitous roles in rewiring the biological pathways. More interestingly, we observed 74.58% of phosSNPs as type III phosSNPs (47, 760), which might suggest that nsSNPs prefer to alter PK types of flanking phosphorylation sites rather than creating or removing phosphorylation sites. Taken together, we proposed that our results could be a useful resource for future disease diagnostics and provide basis for better and individualized. Finally, all phosSNPs data were integrated into PhosSNP 1.0 database, which was implemented in JAVA 1.5 (J2SE 5.0). The PhosSNP 1.0 supports Windows, Unix/Linux and Mac and is freely available for academic researches at: http://phossnp.biocuckoo.org/.
PhosSNP 1.0 User Interface
For publication of results please cite the following article:  PhosSNP for Systematic Analysis of Genetic Polymorphisms That Influence Protein Phosphorylation
PhosSNP for Systematic Analysis of Genetic Polymorphisms That Influence Protein Phosphorylation
Jian Ren, Chunhui Jiang, Xinjiao Gao, Zexian Liu, Zineng Yuan, Changjiang Jin, Longping Wen, Zhaolei Zhang, Yu Xue and Xuebiao Yao. Mol Cell Proteomics. 2010;9(4):623-634 |



